About ERANET-PLL
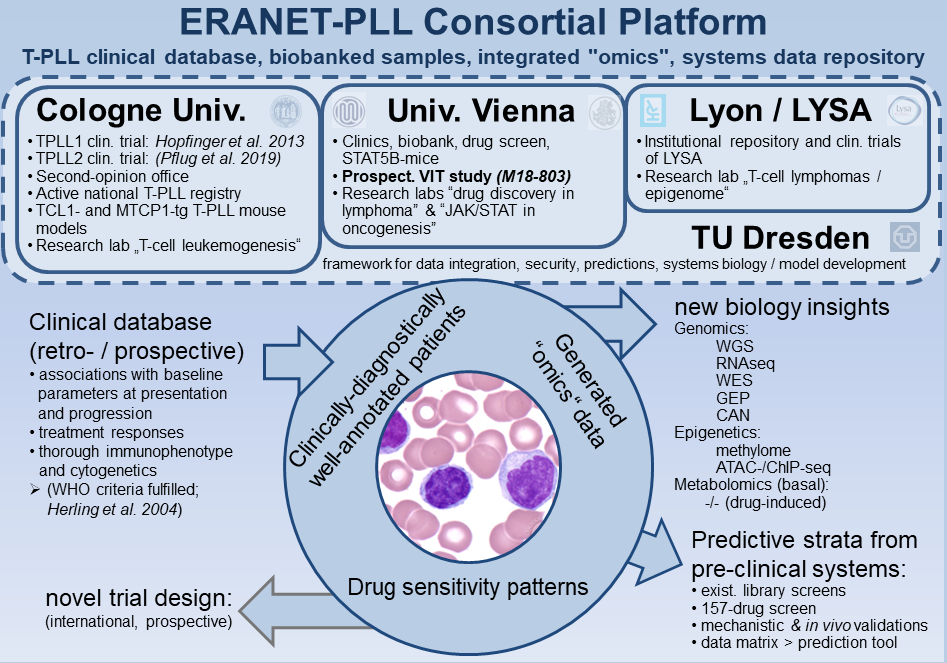
ERANET-PLL is a European consortium of five research groups, with expertise in immunology, clinical hematology, biology, and computational medicine. The aim of this consortium is to implement genomic, epigenetic and metabolic networks in the treatment of T-cell prolymphocytic leukemia (T-PLL), allowing the prediction of most suitable therapies.
Background
T-PLL is the most frequent mature T-cell leukemia, yet it occurs at an incidence of 0.6/Mio in the EU. Its rapid leukemic growth and pronounced chemotherapy resistance translates into an average patient survival of less than 20 months.
Managing T-PLL represents a challenge for even the largest academic centers, as there are no approved drugs available. The CD52-antibody Alemtuzumab, which is used off-label and only made available through a company-supervised named-patient program, is the most active agent. However, response durations rarely exceed 10 months and relapses are the clinical reality for virtually every patient, with hardly any options for a sustained salvage. Further, clinical trials in T-PLL are scarce, mainly due to the impediments by its low frequency and the logistic hurdles imposed by international trials that would be necessary to address drug efficacies in this orphan entity. As a consequence, the median survival of T-PLL patients has not much improved over the last 15 years.
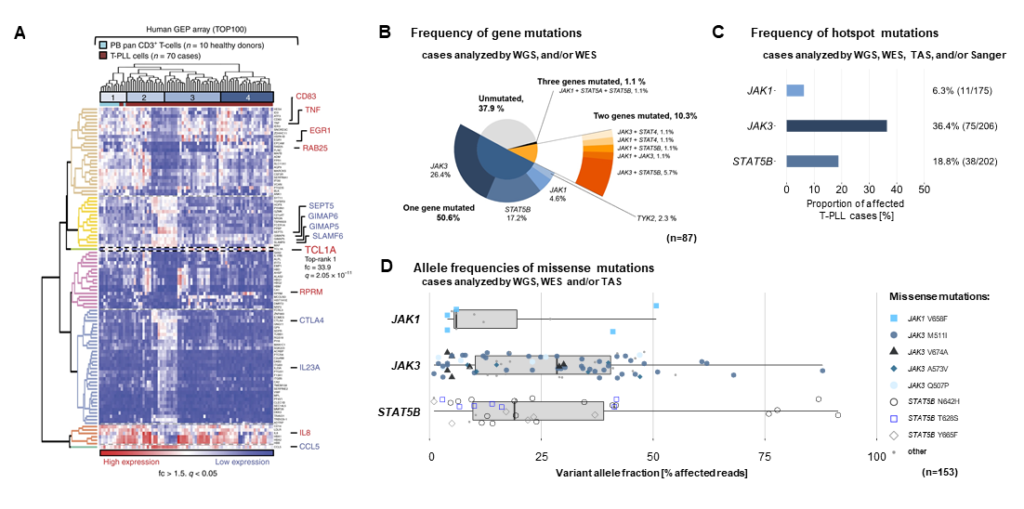
The limited clinical advancements in T-PLL are also rooted in its incomplete biological understanding. Our current molecular concept of T-PLL is based on mostly fragmented and descriptive data. However, large integrative multi-dimensional studies by members of this consortium recently provided deeper insights into the genetic landscape of T-PLL and revealed new vulnerabilities in different pathways. In particular, constitutive expression of the oncogene TCL1, alterations in the DNA damage response (DDR), recurrent hyperactivating mutations in the JAK/STAT pathway, as well as highly frequent lesions in epigenetic regulators were found to be altered in T-PLL. However, epigenetic profiles, i.e. patterns of DNA-methylation, profiles of histone modifications, and states of chromatin accessibility are yet unknown. The metabolic phenotype of T-PLL is also not described beyond a suggested redox-homeostatic imbalance. The consortium will address these knowledge gaps.
Overall, there is an urgent, yet unmet, need for specific and highly efficient drugs that more profoundly eradicate the driving clone(s) in T-PLL. Encouragingly, first compound library screens by groups of this consortium uncovered highly promising vulnerabilities. However, clinically relevant sensitivity clusters of T-PLL patients remain to be identified by a coordinated effort to overcome test platform heterogeneity, incomplete genetic information, limited sample numbers, and paucity of in vivo data.
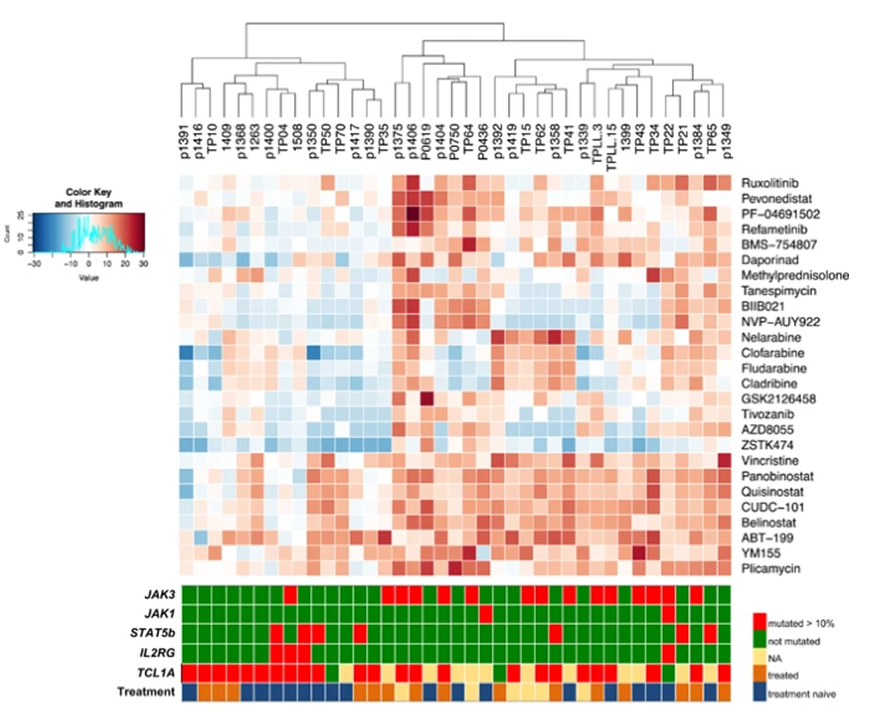
The five teams of ERANET-PLL will capitalize on their unique prerequisites in order to analyze to which degree genomic and epigenetic alterations as well as basal and inhibitor-induced metabolic signatures dictate differential substance activities. Bio-computational modelling will integrate these genotypic and phenotypic dimensions towards prediction tools of in vitro drug sensitivities and synergies. Further, drug candidates will be validated in various preclinical systems. Finally, the extracted set of molecular strata will finally be interrogated in a prospective interventional study.
Aims
Aim I: Establish an advanced lesional map of T-PLL by charting distinctive aberrations in the genomic, epigenetic, and metabolome landscapes of well-characterized cases.
Expanding on an already established large genome and transcriptome data pool, we plan to comprehensively characterize our cohort of T-PLL patients towards a core set of ≈100 cases by supplementing data dimensions to accomplish (near-)complete sample congruence. This well annotated core set will include, among others, data generated by/of:
- Whole genome sequencing (WGS)
- RNA sequencing
- DNA methylation profiles
- ATACseq
- ChiPseq for selected transcription factors
- basal metabolic signatures in sera of T-PLL patients
- metabolite changes in response to (pre)selected inhibitors
Aim II: Identify intra-tumoral and cross-individual patterns of vulnerabilities from a comprehensive drug response chart of T-PLL and validate most specific and active compounds and their combinations.
We aim to conduct compound screens to discern unbiased global drug sensitivity patterns in the clinically and molecularly (Aim I) well-annotated T-PLL core set, using a library that was pre-selected based on our joined preliminary data. In addition, potential synergistic interactions between ≈10 most promising drug candidates at non-toxic dose levels will be assessed in 30 additional T-PLL by combinatorial titrations.
We will then validate candidates in T-PLL samples with defined molecular features, evaluating target pathway inhibition / activation. Most promising candidates will be tested in co-cultures of primary T-PLL cells with stromal cell lines. Having arrived at ≈3 candidate compound (-combinations) with robust in vitro performance, we will expand the experiments in established in vivo systems of available syngeneic murine T-PLL models.
Aim III: Develop a prediction tool of drug sensitivities based on integration of clinical with omics data to stratify for vulnerable patient subsets and most suitable therapies.
We strive to:
- identify inherent molecular patterns that dictate differential in vitro drug responses
- determine relevant disease subsets by association with clinical data
- predict a therapy response of an individual patient’s tumor toward a selected compound, based on a multi-parameter tool
- identify a set of predictors of in-man drug sensitivities and test this tool in the prospective VIT study
Integrating the omics data on the 100 index cases, network building will allow us to identify recurrent or differentially employed core deregulated pathways. In silico interrogations of the data from the genomic, epigenetic, and transcriptomic levels enable functional annotations of genes or branches. Identification of metabolic gene clusters will be aligned with the metabolome profiling data. Pattern recognition by correlation of defined in vitro drug sensitivities with the molecular make up of these cases will finally let us identify predictive biomarkers.
Who benefits?
The work of the ERANET-PLL consortium will provide a benefit not only for other T-PLL researchers, but for many different parties. We aim to:
- increase the current molecular knowledge on T-PLL’s pathogenesis and implement it into clinical application
- advance personalized treatment strategies in T-PLL
- inform professionals as well as non-professionals and raise awareness of T-PLL
- provide relevant disease information to:
- patients and patient organizations
- health care providers and practitioners
- other scientists in the field of T-PLL research
- insurance companies
- industry and biotech
- create an extended data pool for further analyses and research projects
- lay the foundation for future scientific networks in this field
- promote our young scientists and provide them the opportunity to acquire experience, techniques, and concepts within an ambitious research program



